StatComp Project 1: Hints
Hello, dear friend, you can consult us at any time if you have any questions, add WeChat: daixieit
StatComp Project 1: Hints
Avoiding long running times
The problem
Collecting values into a data.frame inside a loop can be slow.
A common issue when combining results computed in loops is that using rbind in each step of the loop to add one or a few rows to a large vector or data.frame can be very slow; instead of linear computational cost in the size of the output, the cost can become quadratic; it needs to reallocate memory and copy the previous results, for each new version of the result object.
The solution
The key is to avoid reallocating memory by instead pre-allocating the whole result object before the loop. However there are some subtleties to this, and the examples blow show the differences between approaches.
A common solution is to work with simpler intermediate data structures within the loop, and then collect the results at the end. Indexing into simple vectors is much cheaper than reallocating and copying memory.
Slow:
slow_fun <- function(N) {
result <- data.frame()
for (loop in seq_len(N)) {
result <- rbind(
result,
data.frame(A = cos(loop), B = sin(loop))
)
}
result
}
Better, but even data.frame indexing assignments causes some memory reallocation:
better_fun <- function(N) {
result <- data.frame(A = numeric(N), B = numeric(N))
for (loop in seq_len(N)) {
result[loop, ] <- c(cos(loop), sin(loop))
}
result
}
Fast; several orders of magnitude faster (see timings below) by avoiding memory reallocation inside the loop:
fast_fun <- function(N) {
result_A <- numeric(N)
result_B <- numeric(N)
for (loop in seq_len(N)) {
result_A[loop] <- cos(loop)
result_B[loop] <- sin(loop)
}
data.frame(
A = result_A,
B = result_B
)
}
Not as fast, but a bit more compact; using matrix indexing doesn’t need memory reallocation, but is a bit slower than vector indexing. A potential drawback is that his method only works when all the columns in the output data.frame should be of the same type (numeric), but for cases where it works the code is both simple, readable, and generally quick to run:
ok_fun <- function(N) {
result <- matrix(0, N, 2)
for (loop in seq_len(N)) {
result[loop, ] <- c(cos(loop), sin(loop))
}
colnames(result) <- c("A", "B")
as.data.frame(result)
}
Benchmark timing comparisons:
N <- 10000
bench::mark(
slow = slow_fun(N),
better = better_fun(N),
fast = fast_fun(N),
ok = ok_fun(N)
)
#> Warning: Some expressions had a GC in every iteration; so filtering is disabled.
#> # A tibble: 4 x 6
#> expression min median `itr/sec` mem_alloc `gc/sec`
#> <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
#> 1 slow 2.83s 2.83s 0.354 2.28GB 20.9
#> 2 better 418.19ms 439.24ms 2.28 1.49GB 89.9
#> 3 fast 1.71ms 1.82ms 541. 229.64KB 2.00
#> 4 ok 4.71ms 4.89ms 199. 458.16KB 16.0
Dynamically generated column names
When the size of a matrix or data.frame depends on a parameter, or there are multiple columns with similar names, it can be convenient to dynamically generate column names. The paste() function can be used for this.
For example, if we want a data.frame of size n × m, with column names Name1, Name2, etc, we can use
df <- matrix(runif(15), 5, 3) # Create a matrix with some values in it
colnames(df) <- paste("Name", seq_len(ncol(df)), sep = "")
df <- as.data.frame(df)
The resulting object looks like this:
df
#> Name1 Name2 Name3
#> 1 0.5515520 0.35036987 0.1395648
#> 2 0.9105334 0.22156578 0.7440625
#> 3 0.3381618 0.51634503 0.2999449
#> 4 0.9466709 0.07315431 0.6551299
#> 5 0.7798397 0.44787741 0.7435251
Multi-column summarise constructions
The summarise() method from the dplyr package is usually used to construct simple summaries such as
suppressPackageStartupMessages(library(tidyverse))
df <- data.frame(x = rnorm(100))
df %>%
summarise(
mu = mean(x),
sigma = sd(x)
)
#> mu sigma
#> 1 -0.05458696 0.956979
But what if we have a function that computes both mu and sigma? If we make it return a data.frame, that can be used with summarise:
my_fun <- function(x) {
data.frame(
mu = mean(x),
sigma = sd(x)
)
}
df %>%
summarise(
my_fun(x)
)
#> mu sigma
#> 1 -0.05458696 0.956979
Avoiding unnecessary for-loops
Since most operations and functions in R are vectorised, the code can often be written in a clear way by avoiding unnecessary for-loops. As a simple example, say that we want to compute 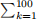 cos(k).
cos(k).
A for-loop solution of the style that would be a good solution in a high performance low-level language like C might look like this:
result <- 0
for (k in seq_len(100)) {
result <- result + cos(k)
}
By taking advantage of vectorised calling of cos(), and the function sum(), we can reformulate into a shorter and more clear vectorised solution:
result <- sum(cos(seq_len(100)))
Other commonly used functions involving vectors of TRUE and FALSE are all() and any(). For example,
vec <- c(TRUE, TRUE, FALSE, TRUE)
all(vec)
#> [1] FALSE
any(vec)
#> [1] TRUE
sum(vec) # FALSE == 0, TRUE == 1
#> [1] 3
Both sum, any, and all also operate across all elements of a matrix. For row-wise and column-wise sums, see rowSums()and colSums().
LaTeX equations
Aligning equations in multi-step derivations
When typesetting multi-step derivations, one should align the equations. In LaTeX, this can be done with the align or align* environments. In some case, that might not work in RMarkdown, but one can then use the aligned environment instead.
Example 1
\begin{align*}
\frac{\partial}{\partial x}f(x, y) &= \frac{\partial}{\partial x}\exp(2x+3y) \\
&= 2\exp(2x+3y)
\end{align*}
Result:
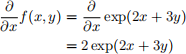
Example 2
$$
\begin{aligned}
\frac{\partial}{\partial x}f(x, y) &= \frac{\partial}{\partial x}\exp(2x+3y) \\
&= 2\exp(2x+3y)
\end{aligned}
$$
Result:
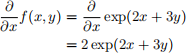
2024-02-27
Avoiding long running times