Midterm exam- BIOS E40 2022
Hello, dear friend, you can consult us at any time if you have any questions, add WeChat: daixieit
Midterm exam- BIOS E40 2022
Question I (37 points)
1
a) 6 pts
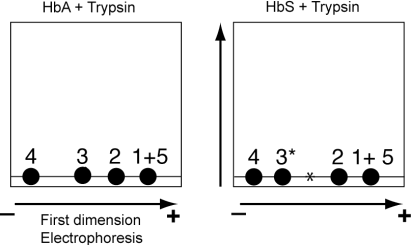
As expected, all the peptides are horizontally aligned in the absence of the second-dimension migration.
The 2D-representation of the peptides indicated that 3 (HbA + trypsin) is further away from 4 than 3* (HbS + Trypsin).
Therefore, panel A represents HbA + Trypsin and panel B represents HbS + Trypsin.
b) 4 pts
In the first-dimension, 1 & 5 are not separated showing that their electrophoretic mobility and most likely their sizes are the same. However, they are separated in the second dimension. Spots 1 and 5 are vertically aligned and must have different hydrophobicity. Since 5 migrates faster than 1, the peptide 5 is more hydrophobic than peptide 1.
c) 6 pts.
The mutation affects the migration of peptide 3 in both dimensions.
Mutant peptide 3 appears less negative (more positive) since its migration is shifted to the left. Therefore, a residue (negatively charged or neutral) is replaced by positively charged residue or a negatively charged residue is replaced by a neutral (or positively charged) residue.
The substitution also increases the hydrophobicity of peptide 3 since mutant peptide 3 migrates faster than wild type peptide 3 in the second dimension.
From both changes one can conclude that after mutation a residue (of unknown charge) is replaced by a residue that is more hydrophobic (and likely neutral, charged residues are hydrophilic).
2
a) 9 pts.
At each cycle, the dominant PTH-amino acid corresponds to the cleaved residue that is to the most N-terminal residue in the processed peptide.
Mother:
Cycle 1: P
Cycle 2: E (note that the peak corresponding to P decreases but do not completely disappear)
Cycle 3: E (the peak E remains very high)
Conclusion:
Mother: 2 HN- __P__ E E -COOH
Same thing for the father:
Father: 2 HN- __P__ __V__ E -COOH
For the child, the second cycle has 2 intense AA-PTH peaks: E and V suggesting that the child has 2 forms of β-hemoglobin, one from the mother (P3 contains E) and one form from the father (P3 contains V).
Therefore:
Child: 2 HN- P _E/V_ E -COOH
b) 4 pts
Peptides are the product of proteolytic digestion using trypsin. Trypsin cleaves after R (arginine) or K (Lysine).
Therefore, the last residue of P3 is either K or R.
c) 4 pts
The disease is recessive, and only homozygous individual (that is the individual should have two mutant copies of the β-hemoglobin gene) will suffer from sickle cell anemia. Heterozygous individuals (1 copy of the mutant gene and 1 copy of the wild type gene) will be carrier of the mutation but will not suffer from sickle cell anemia. Heterozygous individuals will synthesize two different forms of the β-hemoglobin chain (WT protein and mutant protein with 1 amino acid substitution) and would be characterized by two types of peptides 3 (WT and mutant). It appears that the child has 2 types of peptides 3, one WT (...PEE...) inherited from her/his mother and one mutant (...PVE) inherited from her/his father. Therefore, the child will not suffer from the disease.
d) 4 pts.
The mutant β-hemoglobin is characterized by a single amino acid substitution located in peptide 3. The heterozygous child has 2 types of peptides 3 and 6 spots (1,2,3,3*,4, &5) will be visible.
Question II (17 points)
1) 8 pts
X contains the proteins from the H region
Y contains the protein A
The 2 features that dictate this choice are:
- In size exclusion chromatography, large proteins elute before smaller one. Therefore, protein A will elute later than the larger proteins from the H region and A should be found in the second peak.
- The height of the peak is proportional to the amount of protein found in the peak.
Based on the SDS-gel, A is more abundant than the protein from the H region and thus the peak containing A should be higher .
2) 9 pts
S1: F
Explanation: In cation exchange chromatography, the stationary phase is negatively charged and retains positively charged proteins. Therefore, protein A must be positive.
S2: T
Explanation: D represents the migration of the electrophoresis front (bromophenol blue) and d represents the migration of protein A. The ratio d/D can be used in conjunction with a standard curve to calculate the size of A. The standard curve would be built by calculating the d/D ratio for all the protein with a known size present in the ladder.
S3: F
Explanation: SDS would denature proteins and give them an elongated form. The diameter of denatured proteins H and A would increase and as such they apparent size would be larger (denatured and elongated proteins occupy a sphere with a larger diameter than the corresponding native forms). If they appear larger, the peaks would be shifted to the left. It is hard to predict whether the distance between the peaks change.
Question III 12 points
1. 6 pts
Mass accuracy = (Measured mass-monoisotopic mass)/monoisotopic mass
The monoisotopic mass of the protein is 14701 Da
The m/z ratio is 14702.5. The comparison between the m/z ratio and the monoisoptic mass suggest that the ion produced in the instrument carries 1 proton.
We also know that m/z = (Measured mass +1)/1
The measured mass is m/z -1 = 14702.5 -1 = 14701.5
The mass accuracy is (14701.5-14701)/14701 = 3.4 x 10-5 = 34 ppm
2. 6 pts
Based on their position, the difference between a and b represents Δm (the peak width at half the peak height)
Therefore, Δm= 0.125 – (-0.125) = 0.25
The resolution is defined by R= m/Δm = 690.2 /0.25 = 2760.8
Question IV (34 points)
1) 4 pts.
The difference between a b and a c-ion is the additional amino group in the c-ion.
Therefore, the mass (and the m/z ratio) of a c-ions is slightly higher than the mass of a b ion, and the c3 ion will be located on the right side of b3.
2)
The difference in mass between bn (or yn ) and bn+1 (or yn+1 ) is used to identify the residue in position n+1
Ion b1 correspond to A and is not listed in the b-series, therefore the identity of the residue in position 2 cannot be determined from the b-series. Similarly, b8 is unknown and the last residue of the peptide cannot be determined from the b-series.
In position 6 the difference in mass between b5 and b6 correspond to the mass between Leu or Ile (isomeric residues).
a) From the b-ion series (6 pts)
2 HN-A- ? - G - P - K/G - I/L - ? - ? -COOH
Note the first? (Between A and G) can be identified as V: m/z(b1)- (m/z(A) +1)
y1 is known and correspond to arginine (R) + the expected 19Da (OH, H)
y8 is unknown, but we know from the question that the peptide starts with an A
b) From the y-serie (6 pts):
2 HN-A- ? - G - P - K/G - I/L - N - R -COOH
3) 6 pts.
b7 is an ion corresponding to b6 + the residue in position 7 (N):
Therefore m/z (b7) = 566.37 + 114.10 = 680.47
4) 6 pts.
The position of the peak labeled with a black triangle in the spectrum suggest that the corresponding m/z ratio is higher than m/z(b4) and lower than m/z (y3).
From part c) m/z(b7+) = 680.47 (+ indicates the number of associated protons).
Therefore, the mass of b7, M(b7) can be calculated from the formula:
m/z(b7+) = [M(b7) + 1]/1
that is M (b7) = m/z(b7+) – 1 = 679.47
Therefore,
m/z (b7++) = [M(b7) + 2]/2 = 340.73 (m/z ratio for b7 with 2 charges)
m/z (b7+++) = [M(b7) + 3]/3 = 227.49.
Based on the position of the peak labeled with the black triangle you know that the m/z ratio for this peak must be higher than b4 (325.19), therefore the peak labeled with the black triangle correspond to b7 with 2 associated protons.
5) 6 pts
m/z(black triangle) = 340.73 (see calculation above)
Question V (26 points)
1.
a) 4 pts
At the intercept, the net charge of the protein is 0. Therefore, the intercept corresponds to the isoelectric point of the protein .
b) 6 pts.
Anion exchange chromatography is based on the purification of negatively charged proteins (the stationary phase is positively charged). Therefore, to attach to the stationary phase the protein of interest must have net charge that is negative. The isoelectric plot shows that if the buffer has a pH higher than the isoelectric point (5.66) the protein of interest is negatively charged.
c) 6 pts.
A higher pH will be necessary to neutralize the extra basic (positively charged) residues . Therefore, the pI will be higher, and the curve will shift toward the right.
2. 10 pts
The steeper the gradient is the closer the two peaks will be. Therefore, gradient B will reduce the resolution of the two peaks. With gradient A, once the first protein starts to elute from the column (first peak), the salt concentration is maintained constant preventing the elution of the second protein. Then once the first protein is eluted, the salt concentration is increased again to trigger the elution of the 2 proteins. The resolution of the chromatography is improved by delaying the elution of the second protein .
Question VI (28 points)
1)
a) 4pts
The sequence of y5 is C-A-G-A-R-(COOH)
b) 6 pts
The m/z ratio of y5 is 103 + 71 + 57+ 71 +156 + (19) = 477
c) 4 pts
The mass of the peptide before fragmentation is
101 + 71 + 99 + 57 +103 + 71 + 57+ 71 +156 + (1 + 17) = 804 Da
d) 6 pts
The m/z ratio of a peptide with a mass of 804 Da is 805 Da if the peptide carries a single charge. Based on the experimental data, the peptide carries more than one charge since the measured m/z ratio is substantially lower than 805.
We know that m/z = M + (n x 1 Da)/n with M the mass of the peptide, n the number of protons, therefore:
n = M/ [m/z-1] = 804/403-1 = 2
The peptide has 2 charges.
2) 8 pts
The d-series is used to distinguish the 2 isomers Leucine and isoleucine. the d-series is
produced by the cleavage of the β-Y bond on the side chain of the C-terminus residue. if the C- terminus residue is leucine, CH-(C2 H6 ) is removed, and if the C-terminus residue is isoleucine
CH2-CH3 is removed. The mass loss between a b and a d ion is smaller for isoleucine than for leucine.
While the b-ions with Leucine or isoleucine at their C-terminus cannot be distinguished the
derived d-ions have different masses when leucine or isoleucine occupy the C-terminal position.
Question VII (46 points)
1.Completed table (see explanation below) .
Note to the grader: Students don’t have to provide a completed table.

a) 4 pts
The unfragmented peptide contains 8 amino acids. Therefore, y8 and b8 represent the
unfragmented peptide. The question indicates that the m/z ratio for the unfragmented peptide is: 857.545
Therefore m/z(b8) = m/z(y8) = 857.54
b) 4 pts
The following diagram will help you to understand how to calculate the m/z ratio of y3 and other ions .
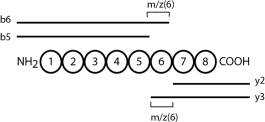
m/z(6) = m/z ratio of the amino acid that occupies position 6
m/z (6) = m/z (b6) – m/z (b5) = m/z (y3)-m/z (y2)
Therefore:
m/z (y3) = m/z (b6) – m/z (b5) + m/z (y2) = 610.39 – 511.32 + 248.16 = 347.23
c) 3 pts
m/z (1) = m/z ratio of the amino acid occupying position 1
m/z (1) = m/z (y8) – m/z (y7)
m/z (1) = 857.54 – 786.50 = 71.04
d) 3 pts
m/z (b1) = m/z (1) + 1 = 71.04 + 1 = 72.04
Why adding 1?
m/z (y8) – m/z (y7) is the m/z ratio of a residue inside a polypeptide chain – [NH-CHR-CO]- see m/z ratio table in the lecture notes .
However, b1 is NH2-CHR-CO, and the extra H adds 1 to the m/z ratio calculated in part c .
e) 4 pts
m/z (2) is the m/z ratio of the amino acid occupying position 2 in the unfragmented peptide
m/z (2) = y7-y6 = 786.5-715.47= 71.03
b2 = b1 + m/z (2) = 143.03
f) 3 pts
m/z (4) is the m/z ratio of the amino acid occupying position 4 in the unfragmented peptide
m/z (4) = y5-y4 545.36 – 446.29 = 99.07
b4 = m/z (4) + b3 = 412.25
g) 4 pts
m/z (Arg) = 156.10
Therefore y1 (Arg) = 156.1 + 19 = 175.1
The 19 da comes from the fact that m/z (arg) read on the table corresponds to the m/z of – [NH- CHR-CO]- , while y1 corresponds to NH2-CHR-COOH + H+
Therefore , you need to consider the added masses of the H (1), the proton (1) and the OH (17): 17 + 1 +1 = 19
m/z (Lys) = 128.09
Therefore y1 (Lys) = 128.09 + 19 = 147.09
h) 4 pts
The m/z ratio of the amino acid occupying position 7 would be:
If the original peptide ends with Arg
m/z (7) = y2 – y1(Arg) = 248.16 – 175.10 = 73.06
If the original peptide end with Lys:
m/z (7) = y2 – y1(Lys) = 248.16 – 147.09 = 101.07
The m/z ratio table indicates that the m/z of 101.07 closely matches one of the numerical values in the table (101.04, Thr) while the m/z (7) of 73.06 does not match any numerical values in the table .
Therefore, the original peptide ends by Lys and y1 = 147.09
i) 2 pts
Based on the answer to the previous question
m/z (7) = y2 – y1 = 248.16 – 147.09 = 101.07
b7 = b6 + m/z (7) = 610.39 + 101.07 = 711.46
2. 4 pts
Now that the table has been completed, the sequence of the peptide can be determined by calculating the difference of m/z ratio between the different ions .
Except for the amino acid labeled X, all the amino acids can be identified based on the difference in m/z ratio between 2 sequential ions .
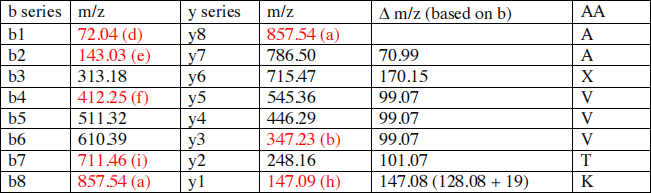
NH2- A-A-X- V-V-V-T-K-COOH
3.
a) 2 pts
1 hydrogen is replaced by CH3-CO therefore the mass shift is the mass of CH3-CO (43) minus the mass of the lost hydrogen (1)
Mass of CH3-CO = (2 x 12) + (3 x 1) + (16) = 43
Mass shift: 43 -1 = 42 Da
b) 4 pts
After deacylation
b3 = 313.18 – 42 = 271.18
y6 = 715.47 – 42 = 673.47
c) 3 pts
The identity of X could be determined by calculating (b3 – b2) or (y6 – y5)
b3 – b2 = 271.18 – 143.03 = 128.15
Based on the m/z ratio table: X = Lys
d) 2 pts
Apparently, the acetylation of Lys, protect this amino acid from trypsin digestion since the acetylated lysine is found inside a tryptic peptide. Most likely, if the deacetylation reaction was performed before the trypsin digestion, the peptide analyzed in this question would not exist and would be replaced by 2 peptides: AAK and VVVTK.
2023-12-19