COMP3419 Graphics and Multimedia Assignment-2 Option-4
Hello, dear friend, you can consult us at any time if you have any questions, add WeChat: daixieit
COMP3419
Graphics and Multimedia
Assignment-2 Option-4
Option 4: Nuclei Segmentation
1 Key Information
• The mark of "COMP3419 Assignment-2 Multimedia Project (Option-4: Nuclei Segmen- tation)" will be given on canvas submission. Due time: 23:59 p.m., Sunday of Week 13 (5-Nov-2023).
• This individual assignment is worth 16% of your final assessment.
• BBBC039 can be downloaded from here. There are also some sample code for evaluation provided on the webstie.
• Submission Deliverable: Students are asked to create a zip file of all deliverables, including a project report (written in LATEX), all source code (with your best model saved), and a demo video. Please be aware of the following submission restrictions that (1) this zip file should be named as "SIDxxxxxxx_Asgmt2Opt4.zip" where xxxxxxx denotes the student ID (e.g., "SID450003419_Asgmt2Opt4"), and (2) the demo video should be named as "SIDxxxxxxx_Asgmt2Opt4.mp4" as well. Failing to follow these restrictions or missing of output video would cause a 5-mark penalty.
• A README txt file to describe the steps/instructions regarding how to get their source code running to derive the expected outputs. This file should help markers get familiar with the submission.
• Submission will only be marked if all deliverables can be accessed from the Canvas System, and they can be runnable following instructions provided in README txt file. Once plagia- rismis detected by the Canvas system, the student will receive zero mark immediately, as well as other related penalties from university.
2 General Marking Policy
Late Submission Policy
For the late submission cases, penalties will be assigned according to the university wide late penalties for assignment Clause 7A of the Assessment Procedures.
Special Consideration and Arrangements
While you are studying, there may be circumstances or essential commitments that impact your academic performance. Our special consideration and special arrangements process is there to support you in these situations. More information on how to lodge the special consideration application, can be found from this webpage.
3 Introduction
Nuclei segmentation is an important task in the realm of biomedical image analysis, playing a fundamental role in various fields such as pathology, cell biology, and medical diagnostics. Through the accurate delineation of cell nucleus boundaries within microscopic images, researchers and clinicians are enabled to extract critical quantitative information about cellular structures, spatial distributions, and morphological characteristics. In this project, you are asked to develop an automated nuclei segmentation system based on learning-based techniques in a well-formulated fluorescence microscopy imaging dataset.
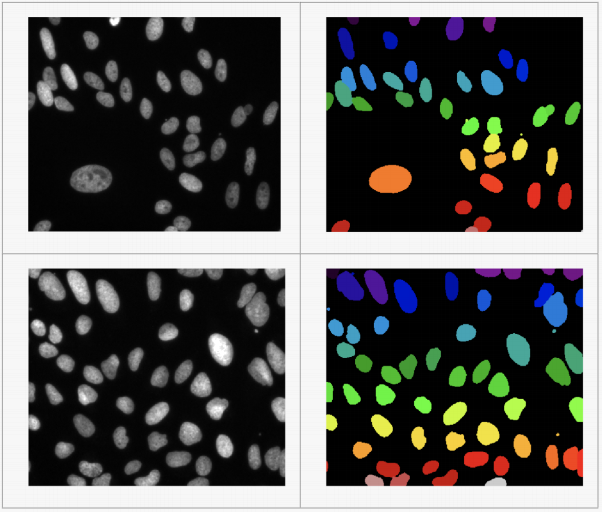
Figure 1: Some examples in the U2OS nuclei dataset.
4 Dataset Description
• The dataset adopted in this assignment is composed of high-throughput chemical screen images on U2OS cells. It comprises a total of 200 fields of view of nuclei captured with fluorescence microscopy using the Hoechst stain. The images are stored as TIFF files with 520x696 pixels at 16 bits. Following the standard procedure in machine learning, we split the dataset into three parts (i.e., training, validation, and test). The detailed file list for each split is provided in the metadata file.
• The ground-truth segmentation masks are provided as PNG files. Note that there will be some cases where more than one nuclei are clustered together. To this end, the mask files label two touching nuclei with different colors for clarification.
• You may need to slice the original 520x696 images into several smaller patches to reduce the hardware requirement, for example, 256x256. Please note that the segmentation masks need to be sliced in the same way to ensure the correct correspondence between the image and mask.
![]() R Hint: To further improve the model performance, some keywords are provided for your own interest: attention network, transformer, generative adversarial network, transfer learning, domain adaptation.
R Hint: To further improve the model performance, some keywords are provided for your own interest: attention network, transformer, generative adversarial network, transfer learning, domain adaptation.
5 Method Design [8%]
4% Students are requested to develop a learning-based automatic segmentation framework for this nuclei segmentation task. Its pipeline might include but not limited to data augmentation, machine learning,deep learning, or a combination of these various types of methods. You can use any open-source libraries, such as Pytorch, scikit-learn, Keras, and Tensorflow.
3% Students are also requested to fine-tune their method, complete performance evaluations (following the evaluation.py given), and discuss the significance of their proposed approach.
1% Some ablation studies should be performed to verify the performance gain caused by each individual component proposed in your approach and/or other attempts made, such as data augmentation techniques adopted, data preprocessing steps implemented and etc.
![]() R You can get some ideas about method design from the research papers uploaded on Canvas (Reference_Papers_Opt4.zip). These papers are well-selected to present a variety of method designs and levels of complexity.
R You can get some ideas about method design from the research papers uploaded on Canvas (Reference_Papers_Opt4.zip). These papers are well-selected to present a variety of method designs and levels of complexity.
6 Project Report [8%]
The report should contain introduction, methods, experimental setup, results & discussion, con- clusion, and references. It should be 2 – 4 pages (maximum 6) in two-column layout, using this template (http://www.latextemplates.com/template/wenneker-article) to follow the sci- entific style and formatted with LATEX1 . A brief guideline of the report sections is as follows:
1% Introduction: introducing the project aim, methods and findings (suggested in 200 - 300 words; no more than 500 words).
1Do not worry if you have not used latex before. It has a similar syntax to HTML. You can find a handy online latex editor at https://www.overleaf.com/.
2% Methods: presenting the details of your method developed, including a brief description of method theories and details in design choices.
1% Experimental setup: describing the dataset and evaluation metrics (suggested in 100 - 200 words; no more than 250 words).
2% Results & Discussion: presenting the evaluation results of each method, including evalua- tion of main design choices, and if applicable presenting results from combining various methods. Based on the experiment results, insightful discussion is expected to demonstrate the corresponding analysis performed. Tables and figures are preferred to be used for result demonstrations.
1% Conclusion: summarising the study and findings (suggested in 100 - 120 words; no more than 150 words).
1% References: listing literature and other references (papers and/or online resources).
7 Deliverable
Students are requested to create a zip file of all deliverables, including
• All the related source code (with your best model saved).
• A PDF project report formatted with LATEX.
• A demo video to introduce your method and findings.
• See Key Information section for submission restrictions.
R Demo Video: The presentation of your method designs, findings, as well as your final results,
should be recorded as a video, using any recording tools available (e.g., Zoom), and the expected
duration is 3-5 mins (no more than 5 mins). It is suggested to use Microsoft PowerPoint/Google
Slides along with the experiment results to present your moddel performance, following the
same structure of the project report.
![]() R Bonus Marks: Different bonus marks will be distributed based on the ranking of the evaluation metrics, over all submissions made to Canvas:
R Bonus Marks: Different bonus marks will be distributed based on the ranking of the evaluation metrics, over all submissions made to Canvas:
• top 10% - 4 marks
• top 50% - 2 marks
• top 85% - 1 mark
Please note that if any flaw or unfair comparisons found in the evaluation results, no bonus mark will be awarded and corresponding marks will be deducted.
8 Google Cloud Platform Education Grant for COMP3419 Students
You can either work locally or use the remote computing platform Google Cloud Engine (click here), which is funded by Google Cloud Platform Education Grant for COMP3419 students, to gain a better computational resources (CPU, GPU,TPU, etc).
Student Google Cloud Coupon Retrieval Access
• The coupon code exclusively for COMP3419 can be redeemedhere. Please keep in mind that there is a redemption limit of 5 coupons per account.
• You will be asked to provide your name and school email address (e.g., [email protected]), which needs to match the domain (please choose the default domain@uni.sydney.edu.au).
An email will be sent to you to confirm these details before a coupon is sent to you.
Watch the Google Cloud Platform Essentials video serieson YouTube.
Instructions for how to redeem grantsin Google Cloud Platform and click "Redeem Now" button.
A comprehensive tutorial on Google Cloud setup and dependencies setup for deep learning- based AI projects can be found from here.
Pleases ensure your instances closed every time when you finish using or training in Google Cloud, to avoid unexpected waste in your coupon.
2023-11-07